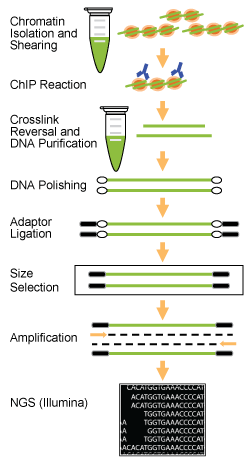
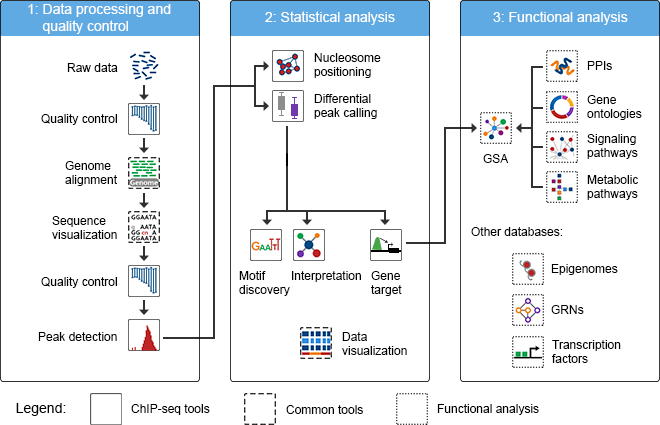
Types of Libraries:
Chromatin immunoprecipitation, or ChIP, is established in epigenetic research as a powerful method for investigating genome-wide DNA-protein interactions in the cell, aiding researchers in understanding chromatin dynamics and epigenetic mechanisms. With the recent technological advancement of next generation sequencing, the ChIP assay can be combined with the power of massively parallel sequencing platforms like Illumina to map the exact binding sites across the entire genome to which the protein of interest is bound.
Sequencing using HiSEQ 2500
Paired End 150 base Read length
40 Million Paired-end Reads
Standard Bioinformatics Included