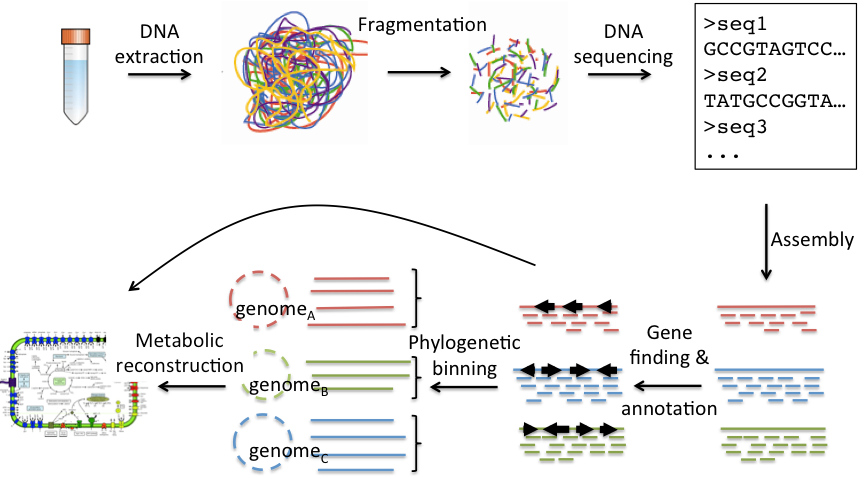
Whole Metagenome Sequencing
Whole Metagenome or Shotgun metagenomic sequencing allows researchers to comprehensively sample all genes in all organisms present in a given complex sample. The method enables microbiologists to evaluate bacterial diversity and detect the abundance of microbes in various environments. Shotgun metagenomics also provides a means to study unculturable microorganisms that are otherwise difficult or impossible to analyze.
Sequencing using HiSEQ 4000
Paired End 150 base Read length
Standard Bioinformatics Included
Standard Whole Metagenome Bioinformatics:
Quality Control
- Raw data QC.
- Removal of low quality reads and trimming of Low quality bases.
- Adaptor trimming.
Whole Metagenome Analysis:
- De-novo assembly of filtered reads.
- Taxonomic classification & Phylogenetic analysis.
- Diversity & Abundance estimation data analysis should be done using optimized and published pipelines.
- Gene catalog generation and expression analysis. Gene annotation.
- Functional GO and Pathway analysis.
- ALPHA & BETA Diversity analysis, 2d & 3D PCA analysis, HEATMAPS, Phylogenetic heat maps, statistical tests, hierarchal clustering etc. ).
- Relative abundance for each gene will be calculated for each individual gene. HEAT MAPS and statistical comparisons in groups.