RRBS- Reduced Representation bisulfite Sequencing
Types of Libraries:
- Chip Sequencing
- Medip Sequencing
- RRBS (Reduced Representation Bisulfite)
- Whole genome (or Oxy) Bisulfite Sequencing
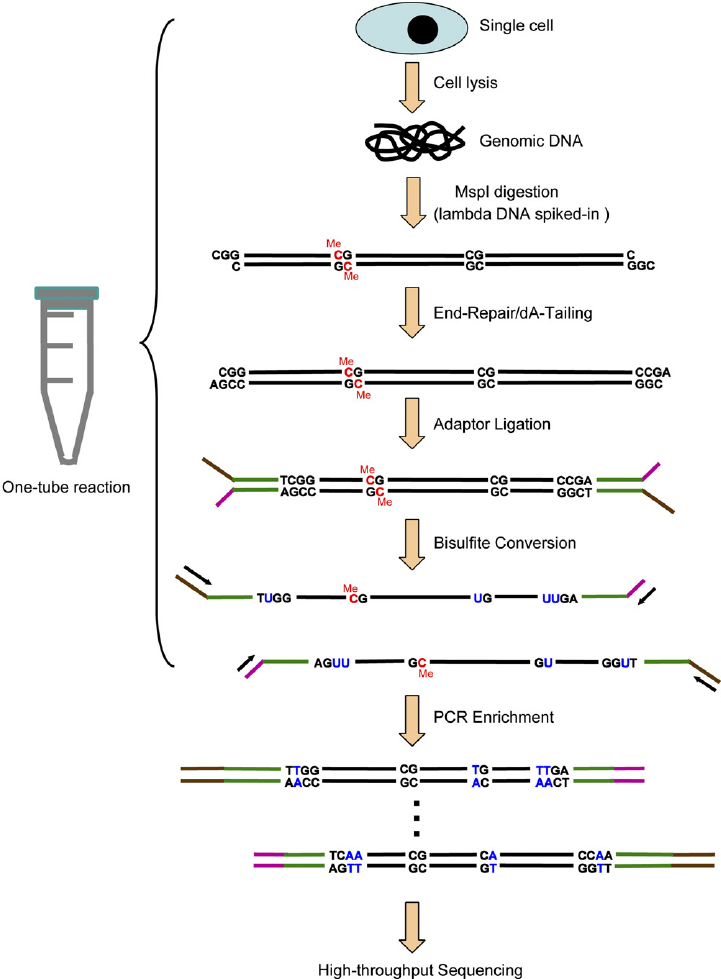
Reduced representation bisulfite sequencing (RRBS) is an efficient and high-throughput technique used to analyze the genome-wide methylation profiles on a single nucleotide level. This technique combines a methylation independent restriction enzyme digest and bisulfite sequencing in order to enrich for the areas of the genome that have a high CpG content. The fragments that comprise the reduced genome include the majority of promotors, as well as regions such as repeat sequences that are difficult to profile using conventional bisulfite sequencing approaches.
Our RRBS service provides our customers with the single-base resolution methylation profile throughout the genome (the Methylome) of their samples.
Highlights of this service:
- Genome-wide
- True single-base resolution
- Applicable to all mammalian samples and tissue types
- Cost-effective technique
Sequencing using HiSEQ 2500
Paired End 150 base Read length
40 Million Paired-end Reads
Standard Bioinformatics Included
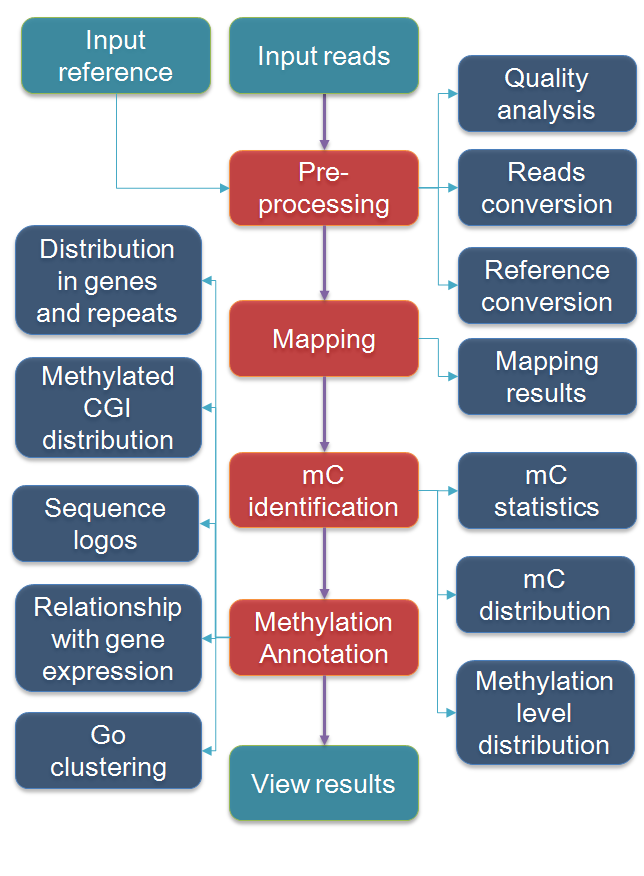
Standard RRBS Bioinformatics:
Quality Control
- Raw data QC.
- Removal of low quality reads and trimming of Low quality bases.
- Adaptor trimming.
RRBS Seq Analysis:
- Mapping of Filtered data to reference.
- Methylated mapped region can be extracted as BED format.
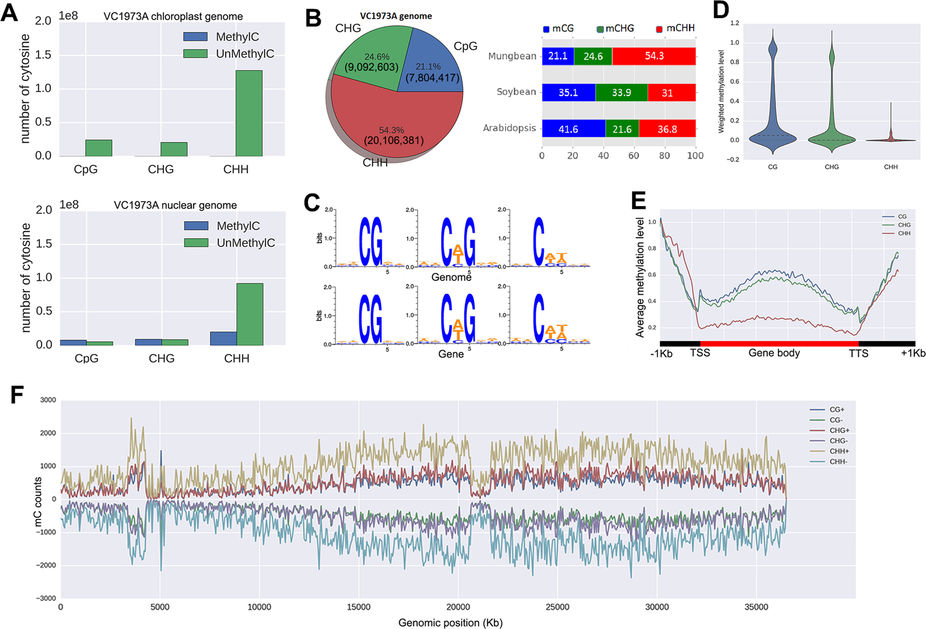
- Genome-wide DNA methylation profile.
- DNA methylation levels in : (i) promoters, (ii) bidirectional promoters, (iii) downstream of a gene and (iv) gene (3′UTRs, 5′UTRs, coding exons and introns)
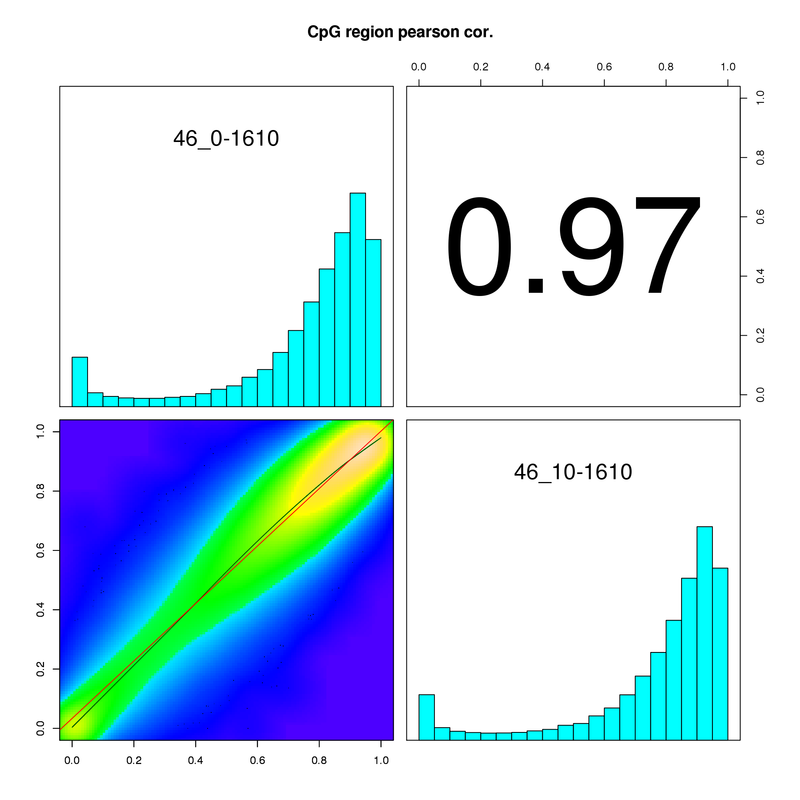
- Differential Methylated regions (DMRs) between samples.
- Gene Target/ Motif identifications.
- Gene Ontology & Pathway Analysis