NGB diagnostics offers multiple RNA Seq (coding & Non Coding) applications portfolio to help you achieve your Expression based project goals for a reasonable price. Whether you are starting with raw material or need help analyzing data produced elsewhere, let our team of experienced scientists help.
We currently offer multiple sequencing platforms including Illumina HiSEQ4000 & X10. We can generate low to high depth genome reequencing for various applications such as:
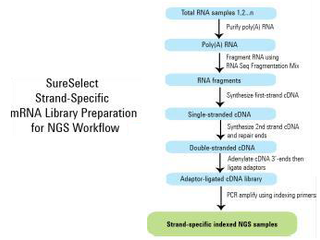
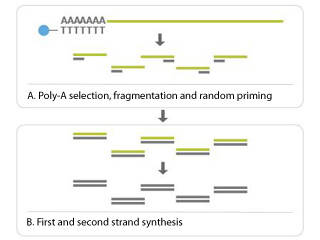
This modification of RNA-Seq is to analyse both Coding and Long Non-Coding (Linc-RNA). The workflow modification is that here from TOTAL RNA, first RIBO depletion is done (NO POLY A enrichment), and straightway taken forward for cDNA conversion and Library preparation.
Sequencing using HiSEQ 4000
2x150 base Read length
40 Million Paired-end Reads
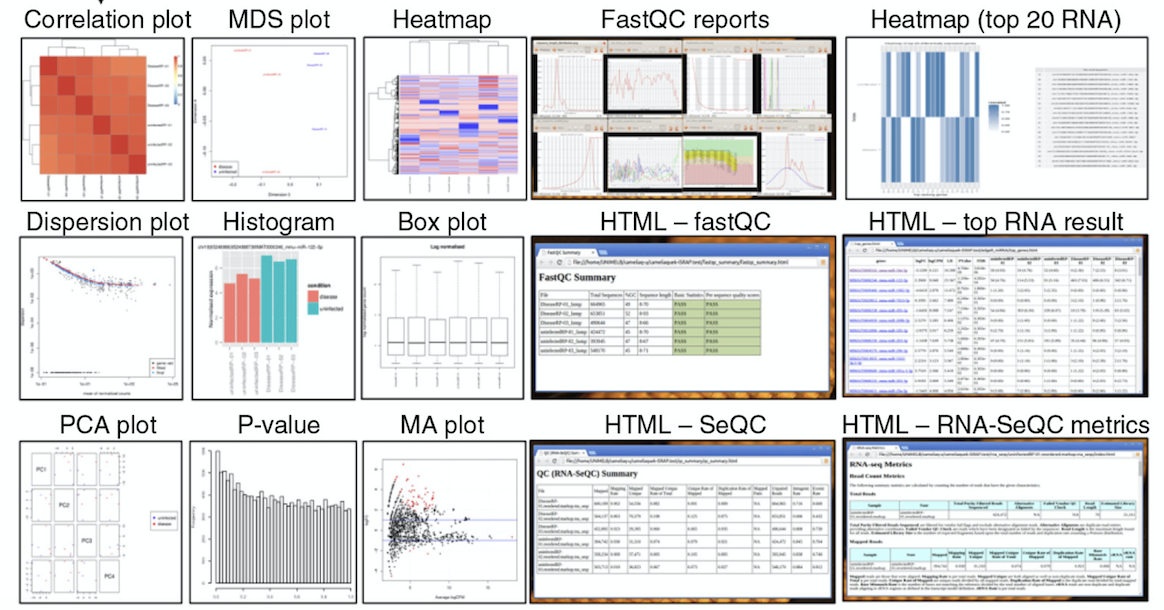
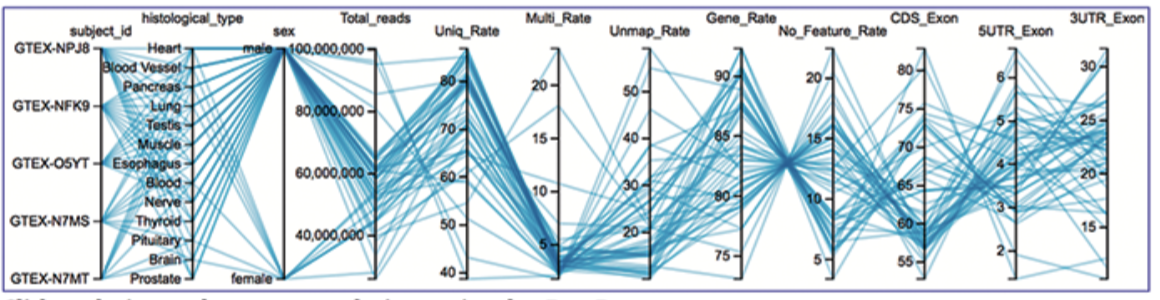
Standard Bioinformatics Included
Sequencing using HiSEQ 4000
2x150 base Read length
40 Million Paired-end Reads
Standard Bioinformatics Included