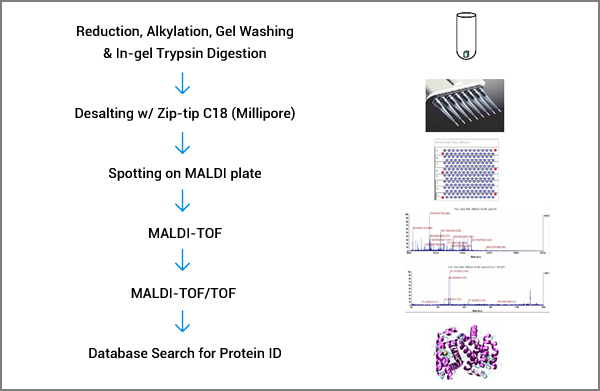
We offer high sensitivity protein identification from gel spots or bands using the latest technologies in mass spectrometry. In each run, four sensitivity standards in the amount of 1 femtomole are included. Our sensitivity is 1 femtomole.
Sample type : well resolved 1D or 2D gel spots containing only 1 protein species