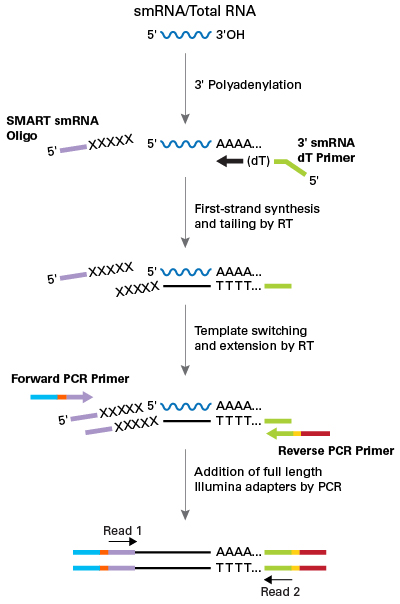
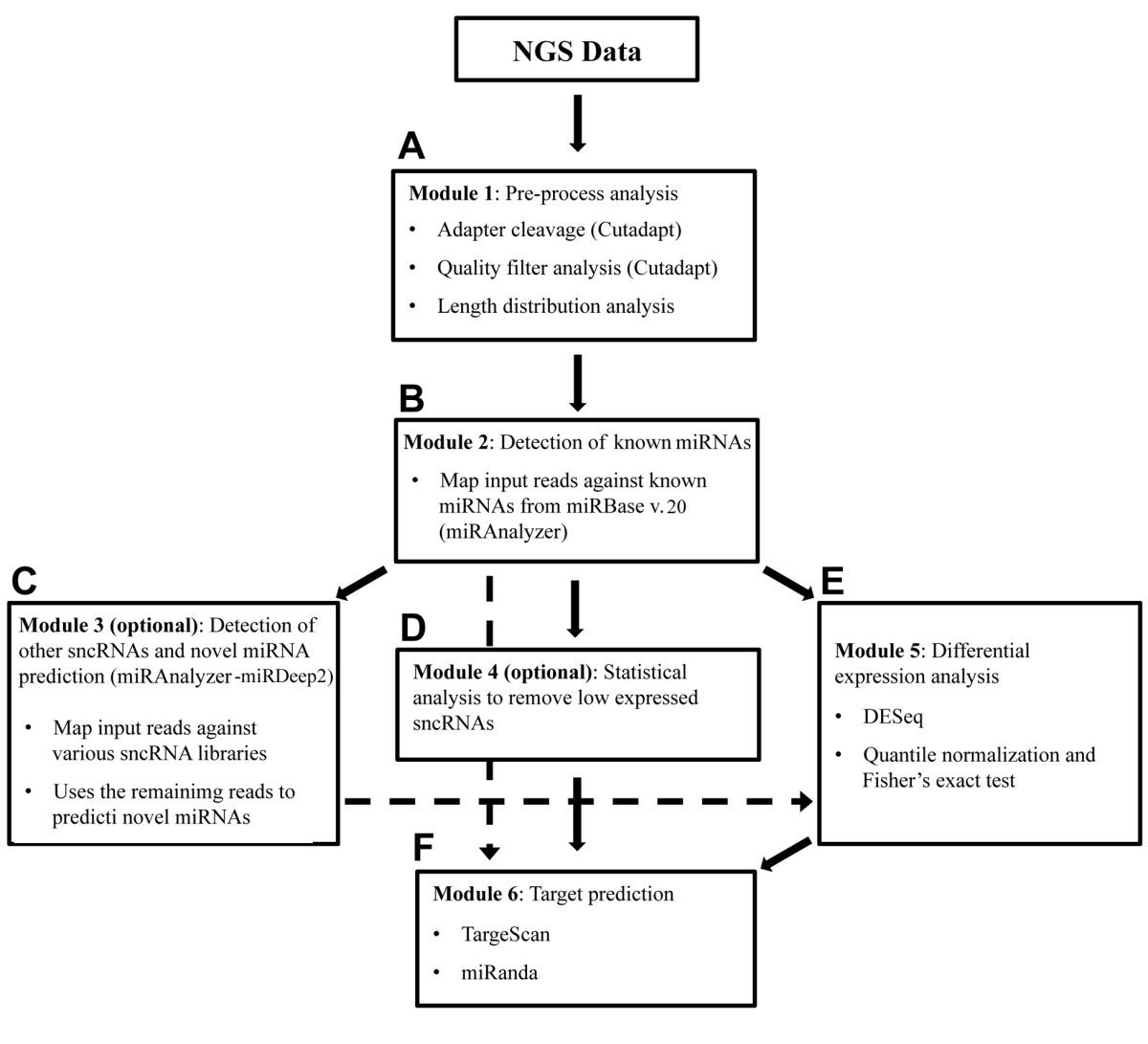
Small RNAs are non-coding RNA (ncRNA) molecule that are less than 200 nucleotides in length are often involved in post-transcriptional regulation through RNA-RNA interactions. Small RNA sequencing involves first isolating then sequencing small RNA species, such as microRNA (miRNA). Small RNA sequencing captures the complete range of small RNA and miRNA species in a sample, and can detect and quantify thousands of small RNA and miRNA sequences with great sensitivity and dynamic range. It has been applied to discover novel miRNAs and other small noncoding RNAs, to examine the differential expression of all small RNAs in and to characeterize variations with single-base resolution.
Sequencing using HiSEQ 2500
Single end 50 base Read length
20 Million Single-end Reads
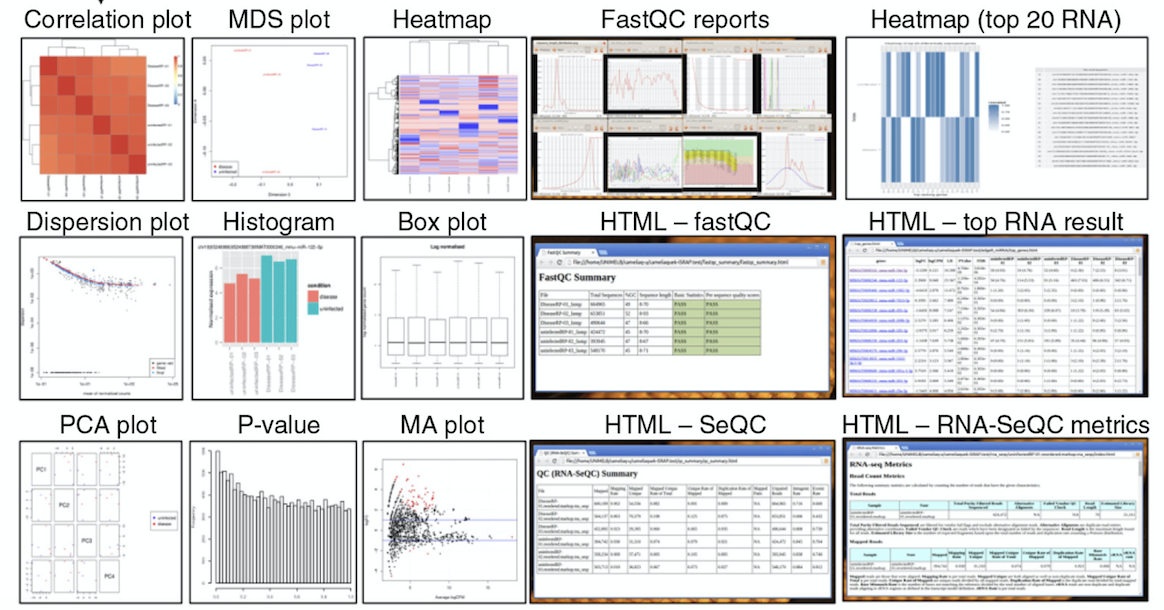
Standard Bioinformatics Included